There are very few inventions over the past 50 years that can compete with the importance of the polymerase chain reaction (PCR) which has fundamentally revolutionised biological and genetic research. PCR is also now key for the diagnosis and treatment of a huge variety of infectious and chronic diseases, including the current outbreak of bird flu.
Since its introduction in 1985, several iterations of the PCR process have been developed and optimised to allow for speedier processing, more nuanced techniques, and a simplified workflow. Now new insulated isothermal PCR (iiPCR) technology has been introduced that makes PCR testing accessible to anyone with minimal training, so enabling rapid diagnostics in any veterinary practice.
The need for rapid diagnostics
From GP vets in front-line clinics to specialist practices, quick and easy diagnostics are essential for providing targeted and appropriate treatment at the point-of-care. Providing the tools for a quicker diagnosis and identification of zoonotic cases means that the correct care pathway can be implemented faster, whether that’s isolation, medication, monitoring or simply advice. This benefits and streamlines the treatment process, minimising risks to veterinary staff and animal owners, eliminating the use of unnecessary drugs, reducing costs for both practices and animal owners, and improving patient recovery times.
This article explores the history and advancements in PCR tools, and discusses how novel insulated isothermal PCR (iiPCR) technology, used within the POCKIT™ Central PCR testing platform (HORIBA, Northampton, UK), significantly simplifies PCR testing making it easy to use in-house. Consequently, staff across a broad range of veterinary settings can now readily and rapidly make confident diagnostic and case management decisions without having to wait for reference laboratory results.
Overview and history of conventional PCR
PCR is a laboratory technique used to make millions of copies of a specified DNA sequence (nucleic acid amplification), making it a powerful tool for highly sensitive and specific pathogen detection. By amplifying a nucleic acid specific to a certain pathogen, this means that PCR tests can detect disease even when there is only a small amount of genetic material present in an original sample.
The fundamental principle behind conventional PCR uses heat and a DNA polymerase enzyme to create the new DNA copies, and its basic process is detailed below.
The conventional PCR process
- Design primers (short DNA sequences) that target a specific sequence of DNA
- Extract the sample DNA
- Make up a tube containing primers, DNA nucleotides, DNA polymerase, and the sample DNA
- Insert this tube into a PCR machine (thermocycler) which will then perform an automatic PCR cycle programme consisting of three steps:
- Denaturing Step: The mixture is heated to about 95°C to separate the double stranded DNA into single strands
- Annealing Step: The mixture is cooled to about 60°C to allow the primers to bind to the target DNA
- Extension Step: The temperature is then increased to approximately 72°C to allow for the DNA polymerase to produce the new strands of DNA starting from the primers
- At the end of the first cycle, each double-stranded DNA molecule consists of one new and one old DNA strand, doubling the previous concentration of that particular sequence
- This cycle is repeated 20-40 times, each time doubling the number of fragments. Using this technique, it is possible to amplify a single piece of DNA to over a billion copies in a few hours
- Finally, use a visualisation technique to see if the DNA sequence being looked for is present (e.g., gel electrophoresis)
Invented by Dr Kary Mullis in 1985, PCR was based on the discovery of a thermostable DNA polymerase (Taq) isolated from the bacterium Thermus aquaticus [1]. This enzyme is capable of surviving prolonged exposure to extreme temperatures as high as 96°C, allowing it to tolerate numerous denaturation cycles. Before this discovery, the polymerase was destroyed in each denaturation step and had to be added for each PCR amplification cycle, making the method tedious and time-consuming.
This discovery of Taq polymerase was revolutionary and resulted in a Nobel Prize in Chemistry in 1993 for Dr Mullis. Since then, major developments in specialised equipment, reagents, sample preparation, computer programs and techniques have been produced to reduce time, error rates and increase the efficiency of PCR.
New technologies – making PCR simple
Despite these improvements in PCR since it was first invented, this technology can still seem technical and arduous to someone who doesn’t work in a lab. Even the basic process described above requires knowledge on primer design, DNA extraction, pipetting techniques, and of methods for visualising the end result. Not only that, but the length of time needed to change from one temperature to another during PCR can be slow and inefficient, leading to reactions taking many hours. It is no wonder there has been a perception that conventional PCR is complex, requiring large and multiple pieces of equipment plus specialist staff.
However, new technologies are being developed all of the time and PCR is no exception to this. Proven insulated isothermal PCR (iiPCR) technology greatly reduces the transition time between temperatures by using the natural temperature gradient generated from thermal convection to drive PCR [2-4] (Figure 1). Essentially, one PCR nucleic acid duplication cycle is completed with every thermal convection cycle within a tube. This eliminates the need for the many cycles of heating and cooling used by conventional PCR, thus enabling significantly quicker diagnostic procedures that are still accurate and reliable.
Furthermore, iiPCR technology not only significantly reduces PCR reaction time, it also means that much smaller equipment can be used. This is because iiPCR requires just a single isothermal heat source of 95°C at the base of special-designed capillary tubes to induce the natural thermal convection that drives the PCR reaction and the temperature gradient in an insulated environment.
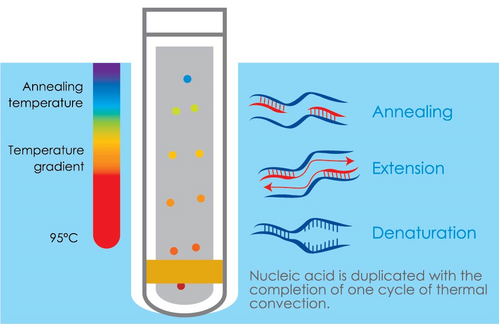
Figure 1: Insulated isothermal PCR (iiPCR) technology explained - unlike conventional PCR, which requires several cycles of heating and cooling, iiPCR is achieved through the temperature gradient generated from thermal convection which enables a significantly reduced reaction time.
Sample-in, result-out process
Incorporating the benefits of simplicity, size and speed enabled by iiPCR technology, HORIBA’s POCKIT™ Central is an extremely easy-to-use, small benchtop PCR amplification and detection system (Figure 2). It can complete molecular diagnostic tests in just 85 minutes, providing rapid and accurate assessment for a wide range of pathogens.

Figure 2: HORIBA’s POCKIT Central in-house PCR testing platform is very compact and simple to use.
In addition to being quick, POCKIT Central automates and optimises every stage of the PCR process to enable walk-away in-house nucleic acid detection. It fully integrates and automates DNA extraction, PCR amplification, and detection in one instrument to offer a fast and easy molecular diagnostic system that can be used with minimal training and no other equipment required.
Quality DNA from multiple samples can be extracted by directly inserting the sample and pressing the “START” button on the user-friendly touch screen where final test results are also displayed. Data is also automatically stored on the analyser which can be exported via USB or WiFi. Additionally, ready-to-use cartridge-based consumables and universal pre-programmed protocols allow users to greatly reduce hands-on time and simplify their workflow.
When using POCKIT Central based on iiPCR technology, users need not know the long list of conventional PCR process stages detailed in the information box and replace them simply with:
- Sample-in
- Result-out
Enabling in-house testing benefits
Currently, most vets seeking the specificity of a PCR test will send samples out to their nominated reference laboratory, involving sample packaging and carriage, awaiting a test run and for results to be reported. Moving more diagnostic tests in-house vastly reduces turnaround times, enabling vets to make informed decisions on patient management, quickly administer effective treatment or consider further tests, if necessary. In many cases, iiPCR used in practice can provide a rapid and accurate test for infectious pathogens to help reduce disease spread among companion and livestock animals.
For example, due to increasing numbers of imported dogs in the UK, there have been more reports of exotic parasites and diseases. These include tick-borne pathogens like Ehrlichia canis, Anaplasma platys, Hepatozoon canis and Babesia spp, and other parasites such as Brucella canis and Dirofilaria immitis. Since many infectious diseases can remain silent and only become apparent months to years after infection, it’s important that diseases can be diagnosed soon after travel or importation to prevent transmission and serious illness further down the line. Using simple in-house iiPCR in the form of HORIBA’s POCKIT Central to test for such infections provides highly accurate and specific results, allowing for targeted and effective treatment (5).
Offering over 190 assays tailored towards small animals, aquaculture, livestock and equine pathogens, POCKIT Central can run up to 8 samples simultaneously for single or multiple pathogens with no risk of cross-contamination in just 85 minutes. This is a huge reduction in time when compared to the process of sending samples to large-scale, commercial labs which can take anything from 24 hours to 10 days to deliver results, due to transportation or batching of tests. Early detection and diagnosis in-house can improve workflow, reduce the risk of further infection or relapse and subsequent trips to the veterinary clinic, lowering costs for both practices and owners while improving patient recovery times.
POCKIT Central has a footprint of just 31 x 48cm and is extremely easy to set up and operate, allowing a laboratory to potentially be operational and offering PCR-based diagnostic tests within 30 minutes. Tests can be run by any trained staff, eliminating the need for specialised personnel, and allowing for consistent contact and management for each patient. POCKIT Central is a cost effective and simple way for veterinary practices to increase diagnostic capacity, reduce waiting times, and enhance patient care by delivering best practice, evidence-based treatment.
In summary
Due to the proven iiPCR-based technology within HORIBA’s POCKIT Central, PCR-based testing can now be very easily and effectively used in-house in any veterinary practice as a simple to use tool enabling rapid, cost-effective diagnostics for a variety of diseases. As this article demonstrates, new iiPCR technology might be clever, but its implementation is very simple and the benefits of use are great!
References:
- Saiki, R.K. et al. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science (New York, N.Y.) 239(4839), pp. 487–491. doi: 10.1126/science.2448875.
- Krishnan, M., Ugaz, V.M. and Burns, M.A. 2002. PCR in a Rayleigh-Bénard Convection Cell. Science. Available at: www.science.org/doi/10.1126/science.298.5594.793
- Chou, W.P., Chen, P.H., Miao, M., Kuo, L.S., Yeh, S.H. et al. 2011. Rapid DNA amplification in a capillary tube by natural convection with a single isothermal heater. BioTechniques 50(1), pp. 52–57. doi: 10.2144/000113589.
- Tsai, Y.-L. et al. 2012. Development of TaqMan probe-based insulated isothermal PCR (iiPCR) for sensitive and specific on-site pathogen detection. PloS One 7(9), p. e45278. doi: 10.1371/journal.pone.0045278.
- Martin, L. 2022. Imported pets – bringing in more than you bargained for? Veterinary Practice. Available at: www.veterinary-practice.com/article/parasites-in-imported-pets